6 Processing many polar volumes
6.1 Accessing radar data
- US NEXRAD polar volume data can be accessed in the Amazon cloud. Use function
download_pvolfiles()for local downloads - European radar data can be accessed at https://aloftdata.eu. These are processed vertical profiles, the full polar volume data are not openly available for most countries. Data are available for single profiles (vp) in ODIM h5 format, or as monthly vertical profile time series (vpts) in VPTS CSV format.
- Use function
download_vpfiles()for local vp downloads, or manually download monthly vpts files that can be read withread_vpts().
The names of the radars in the networks can be found here:
- US: NEXRAD network
- Europe: OPERA database
Useful sites for inspecting pre-made movies of the US composite are https://birdcast.info/migration-tools/live-migration-maps/ and https://www.pauljhurtado.com/US_Composite_Radar/.
6.2 Processing multiple polar volumes
This section contains an example for processing a directory of polar volumes into profiles:
First we download more files, and prepare an output directory for storing the processed profiles:
# First we download more data, for a total of one additional hour for the same radar:
download_pvolfiles(date_min=as.POSIXct("2017-05-04 01:40:00"), date_max=as.POSIXct("2017-05-04 02:40:00"), radar="KHGX", directory="./data_pvol")## Downloading data from noaa-nexrad-level2 for radar KHGX spanning over 1 days##
## Downloading pvol for 2017/05/04/KHGX/# We will process all the polar volume files downloaded so far:
my_pvolfiles <- list.files("./data_pvol", recursive = TRUE, full.names = TRUE, pattern="KHGX")
my_pvolfiles
# create output directory for processed profiles
outputdir <- "./data_vp"
dir.create(outputdir)We will use the following custom settings for processing: * We will use MistNet to screen out precipitation * we will calculate profile layers of 50 meter width * we will calculate 60 profile layers, so up to 5060=3000 meter altitude we will store the profiles in VPTS CSV format, by specifying an output file with extension .csv (this file format is fast for reading data back in later)
Note that we enclose the calculate_vp() function in tryCatch() to keep going after errors with specific files
Having generated the profiles, we can read them into R:
# we assume outputdir contains the path to the directory with processed profiles
my_vpfiles <- list.files(outputdir, full.names = TRUE, pattern="KHGX")
# print them
my_vpfiles## [1] "./data_vp/KHGX20170504_012843_V06_vp.csv"
## [2] "./data_vp/KHGX20170504_013405_V06_vp.csv"
## [3] "./data_vp/KHGX20170504_013934_V06_vp.csv"
## [4] "./data_vp/KHGX20170504_014502_V06_vp.csv"
## [5] "./data_vp/KHGX20170504_015031_V06_vp.csv"
## [6] "./data_vp/KHGX20170504_015600_V06_vp.csv"
## [7] "./data_vp/KHGX20170504_020127_V06_vp.csv"
## [8] "./data_vp/KHGX20170504_020656_V06_vp.csv"
## [9] "./data_vp/KHGX20170504_021224_V06_vp.csv"
## [10] "./data_vp/KHGX20170504_021752_V06_vp.csv"
## [11] "./data_vp/KHGX20170504_022320_V06_vp.csv"
## [12] "./data_vp/KHGX20170504_022848_V06_vp.csv"
## [13] "./data_vp/KHGX20170504_023417_V06_vp.csv"
## [14] "./data_vp/KHGX20170504_023946_V06_vp.csv"# read them into a vpts object:
my_vpts <- read_vpts(my_vpfiles)You can now continue with visualizing and post-processing as we did earlier:
# plot them between 0 - 3 km altitude:
plot(my_vpts, ylim = c(0, 3000))## Warning in plot.vpts(my_vpts, ylim = c(0, 3000)): Irregular time-series:
## missing profiles will not be visible. Use 'regularize_vpts' to make time series
## regular.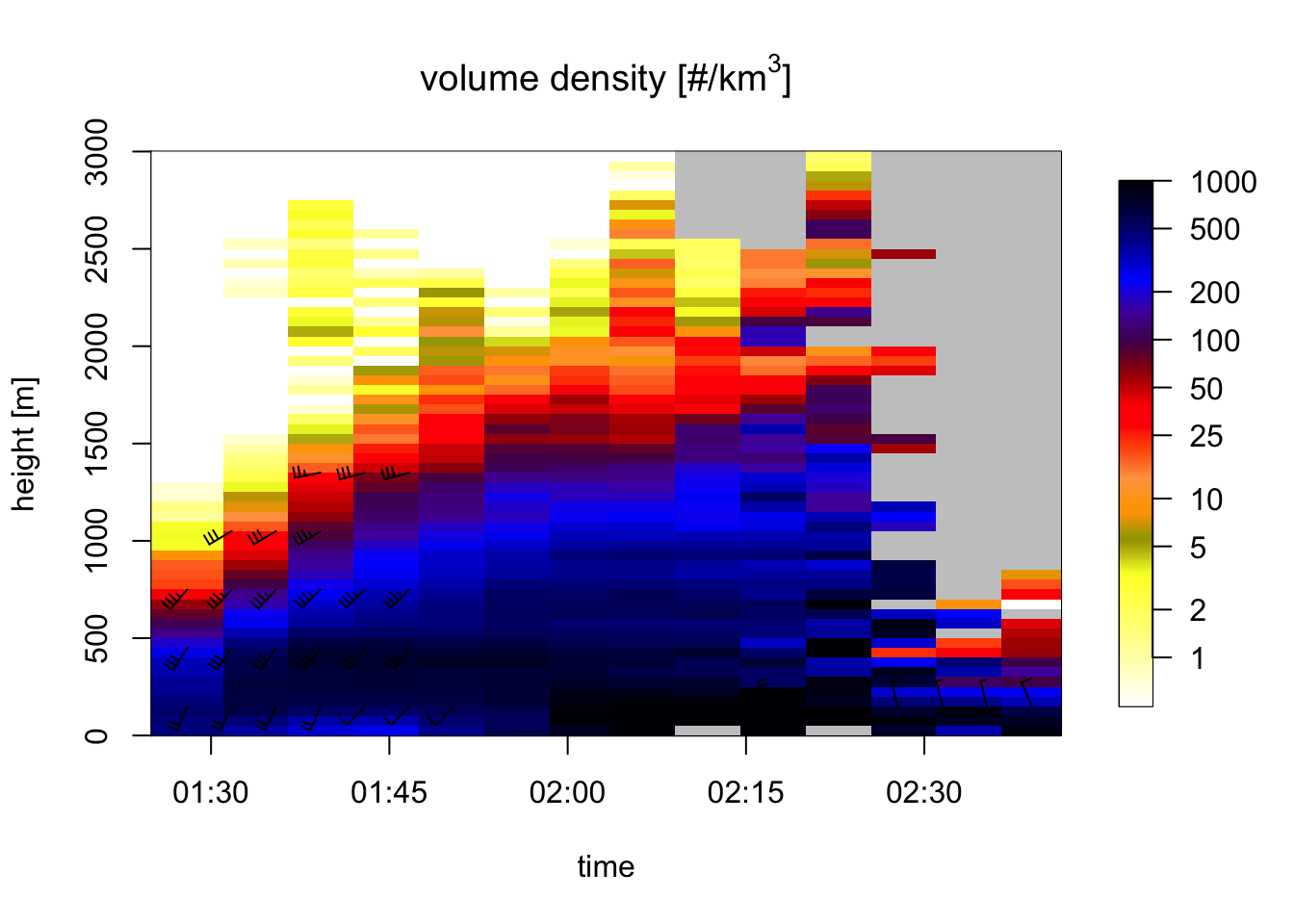
# because of the rain moving in, our ability to estimate bird profiles slowly drops:
# let's visualize rain by plotting all aerial reflectivity:
plot(my_vpts, ylim = c(0, 3000), quantity="DBZH")## Warning in plot.vpts(my_vpts, ylim = c(0, 3000), quantity = "DBZH"): Irregular
## time-series: missing profiles will not be visible. Use 'regularize_vpts' to
## make time series regular.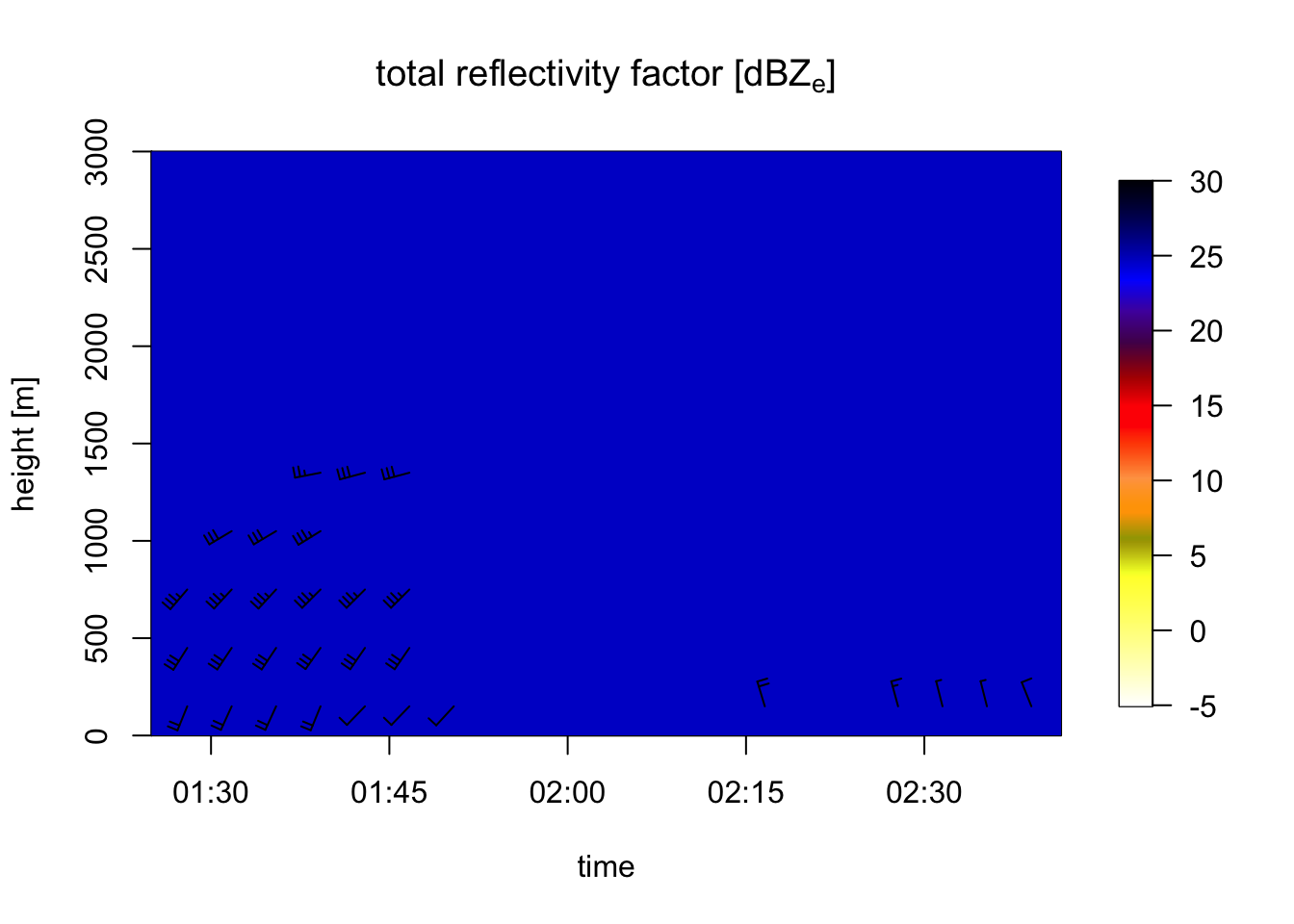 Note that we enclose the
Note that we enclose the calculate_vp() function in tryCatch() to keep going after errors with specific files
6.3 Parallel processing
We may use one of the parallelization packages in R to further speed up our processing. We will use mclapply() from package parallel. First we wrap up the processing statements in a function, so we can make a single call to a single file. We will disable MistNet, as this deep-learning model does not parallelize well on a cpu machine.
process_file <- function(file_in){
# construct output filename from input filename
file_out <- paste(outputdir, "/", basename(file_in), "_vp.csv", sep = "")
# run calculate_vp()
vp <- tryCatch(calculate_vp(file_in, file_out, mistnet = FALSE, h_layer=50, n_layer=60), error = function(e) NULL)
if(is.vp(vp)){
# return TRUE if we calculated a profile
return(TRUE)
} else{
# return FALSE if we did not
return(FALSE)
}
}
# To process a file, we can now run
process_file(my_pvolfiles[1])## Filename = ./data_pvol/2017/05/04/KHGX/KHGX20170504_012843_V06, callid = KHGX
## Removed 1 SAILS sweep.
## Call RSL_keep_sails() before RSL_anyformat_to_radar() to keep SAILS sweeps.
## Reading RSL polar volume with nominal time 20170504-012844, source: RAD:KHGX,PLC:HOUSTON,state:TX,radar_name:KHGX
## Running vol2birdSetUp
## Warning: disabling single-polarization precipitation filter for S-band data, continuing in DUAL polarization mode## [1] TRUENext, we use mclapply() to do the parallel processing for all files:
# load the parallel library
library(parallel)
# detect how many cores we can use. We will keep 2 cores for other tasks and use the rest for processing.
number_of_cores = detectCores() - 2
# Available cores:
number_of_cores
# let's loop over the files and generate profiles
start=Sys.time()
mclapply(my_pvolfiles, process_file, mc.cores = number_of_cores)
end=Sys.time()
# calculate the processing time that has passed:
end-start